Session metadata tutorial
This tutorial shows you how to generate the session metadata struct used by CellExplorer. 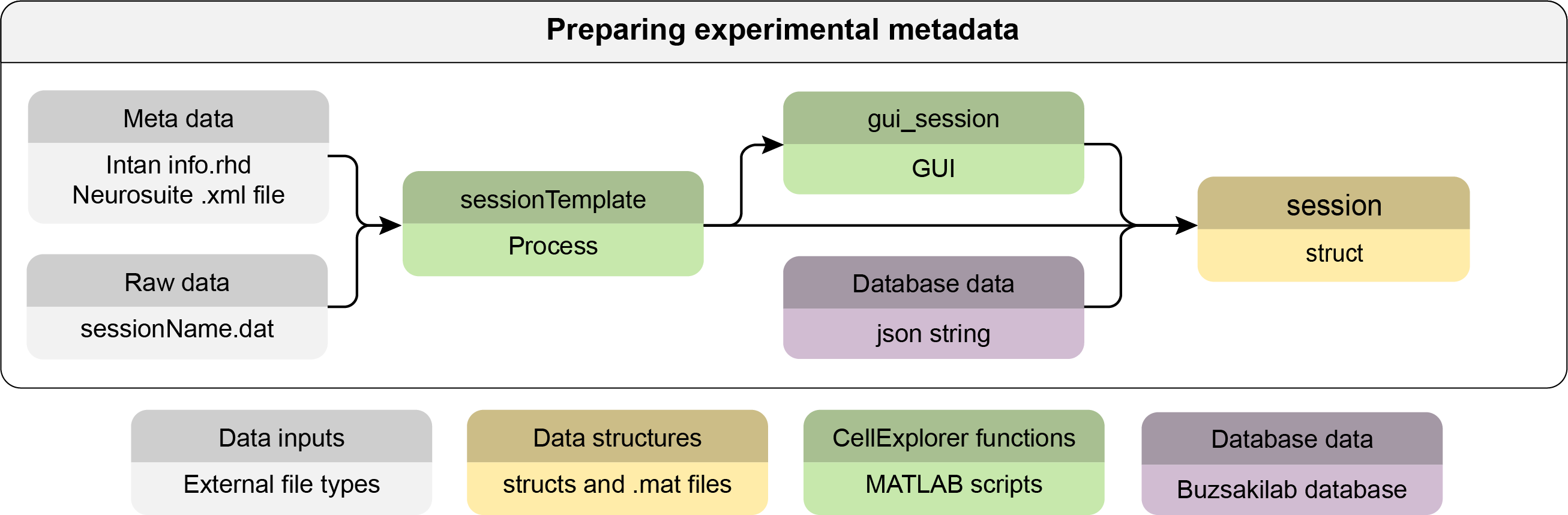
- Define the basepath of the dataset to run. A valid dataset should consist of a binary raw data file and spike sorted data.
basepath = '/your/data/path/basename/'; cd(basepath)CellExplorer operates with one main path for a dataset: the
basepathwhich defines the local path to the dataset. The session name, also referred to asbasename, is assumed to be the same as the directory of the session. These fields are defined in thesession.generalstruct. The raw data file should be located in thebasepath. - You can generate the session metadata struct using the session template script
session = sessionTemplate(basepath);sessionTemplatewill extract metadata from the content ofbasepath. Please go through thesessionTemplatescript to understand the various components that are extracted. You can create your own template scripts from the original, adding any relevant metadata. - Use the session gui for inspecting the metadata struct and for further manual entry.
session = gui_session(session);Below is a screenshot of the metadata interface with metadata entered:
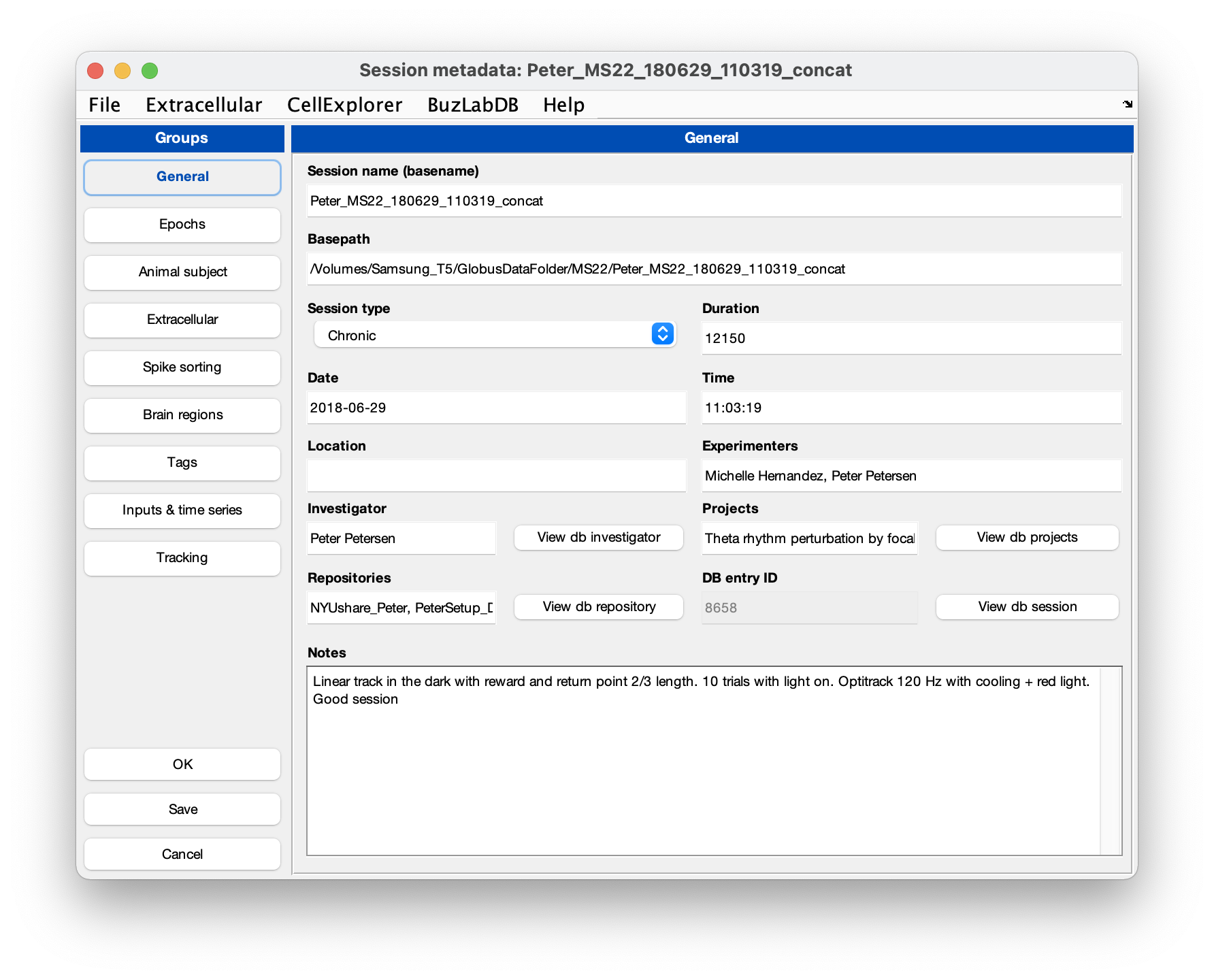
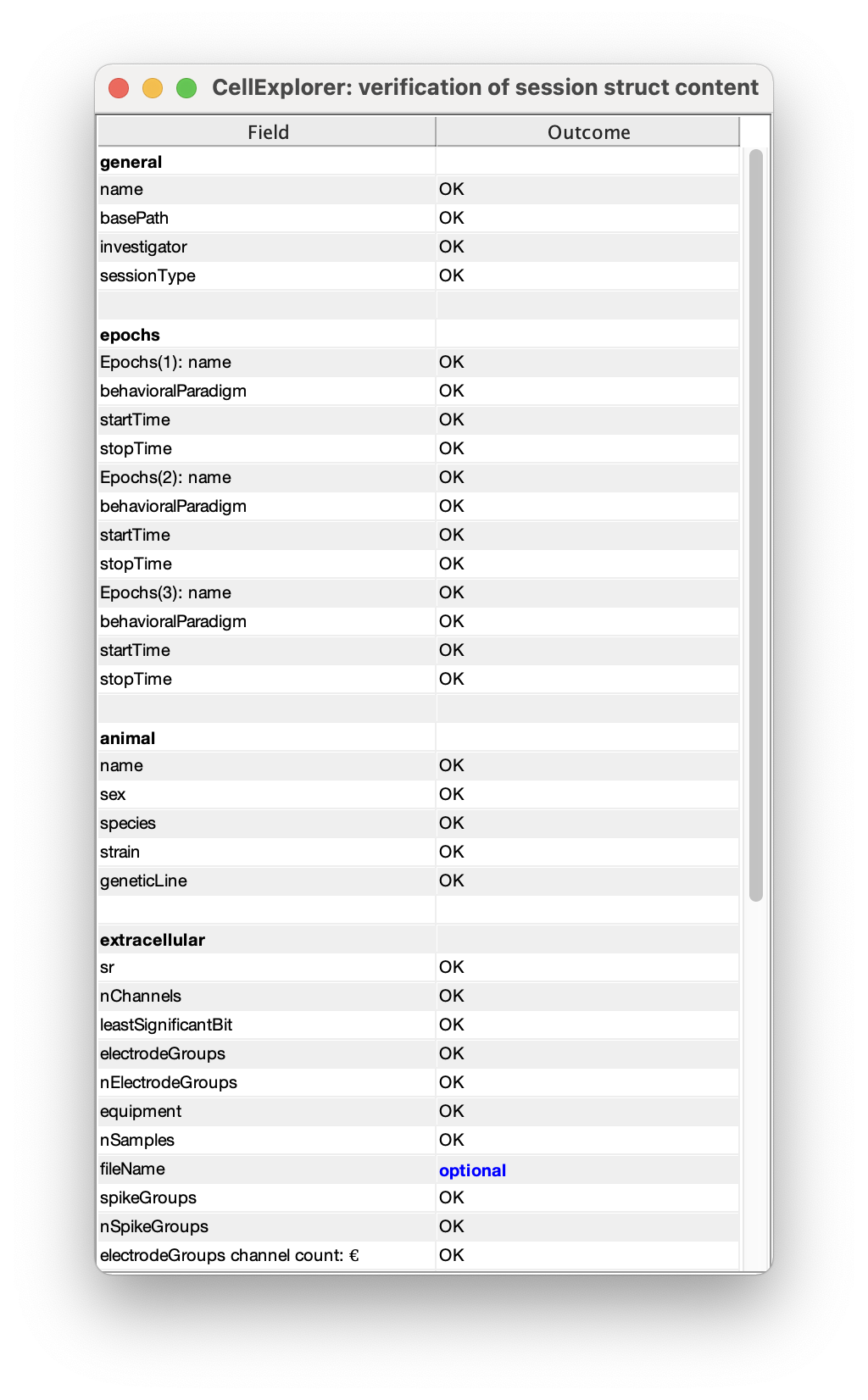
- You can validate the entered metadata by running the verification script:
validateSessionStruct(session);This will show a table, with missing required fields highlighted in red, and unused optional fields in blue. The verification can also be run from the session GUI: