Framework for analyzing single cells
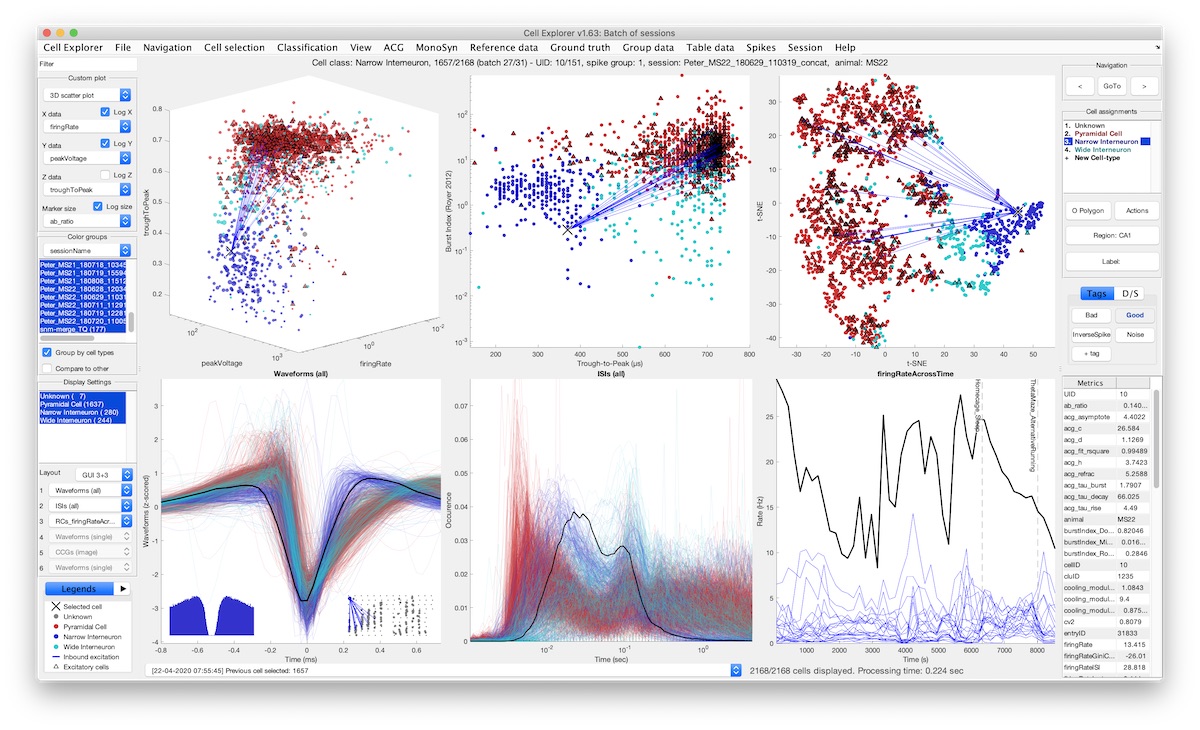
CellExplorer is a graphical user interface (GUI), a standardized processing module and data structure for exploring and classifying single cells acquired using extracellular electrodes.
Get started now View code on GitHub
Introduction
The large diversity of cell-types of the brain, provides the means by which circuits perform complex operations. Understanding such diversity is one of the key challenges of modern neuroscience. Neurons have many unique electrophysiological and behavioral features from which parallel cell-type classification can be inferred.
To address this, we built CellExplorer, a framework for analyzing and characterizing single cells recorded using extracellular electrodes. It can be separated into three components: a standardized yet flexible data structure, a single yet extensive processing module, and a powerful graphical interface. Through the processing module, a high dimensional representation is built from electrophysiological and functional features including the spike waveform, spiking statistics, monosynaptic connections, and behavioral spiking dynamics. The user-friendly interactive graphical interface allows for classification and exploration of those features, through a rich set of built-in plots, interaction modes, cell grouping, and filters. Powerful figures can be created for publications. Opto-tagged cells and public access to reference data have been incorporated to help you characterize your data better. The framework is built entirely in MATLAB making it fast and intuitive to implement and incorporate CellExplorer into your pipelines and analysis scripts. You can expand it with your metrics, plots, and opto-tagged data. The paper is published in Neuron: https://doi.org/10.1016/j.neuron.2021.09.002.
Data structure Processing module Graphical interfaces
Getting started
- Clone, fork, or download the repository (cloning or forking is recommended).
- Add the local repository to your MATLAB setpath (make sure to select Add with subfolders…).
- The pipeline uses two c-code files that must be compiled
CCGHeart.candFindInInterval.c(originally part of the FMA toolbox). Compiled versions are included for Windows 64bit, Intel Mac 64bit, and Apple Silicon (ARM; M1 and later), but you have to compile the scripts if your OS version is different (e.g. Linux). In MATLAB, go toCellExplorer/calc_CellMetrics/mex/and run these line:mex -O CCGHeart.c mex -O FindInInterval.c - CellExplorer uses additional MATLAB toolboxes, where three of them are required.
- Curve Fitting Toolbox (required).
- Signal Processing Toolbox (required).
- Statistics and Machine Learning Toolbox (required).
- Parallel Computing Toolbox (optional, allows for parallel processing of certain features).
- Image Processing Toolbox (optional for NeuroScope2, allows for selection of channels from the probe layout).
- DSP System Toolbox or the Audio Toolbox (optional for NeuroScope2 to stream audio of traces).
That’s it! Now you can explore the software with below example data or try one of the tutorials with your own data.
Try CellExplorer with example data
There is an example dataset included in the repository for trying CellExplorer. Load the mat-file cell_metrics_batch.mat into MATLAB and type:
CellExplorer('metrics',cell_metrics)
Tutorials for using the framework with your own data
We have created a few tutorials to get you started, covering the pipeline and the graphical interface. There is also a tutorial script: CellExplorer_Tutorial.m included with example code for running the pipeline and the GUI on your data.
Please also check out the recorded presentation of CellExplorer at the Neuropixels course in 2022. It includes an introduction with slides and a demo in Matlab showing how to use the processing pipeline, CellExplorer, and NeuroScope2 (11 minutes long).
Support
Please use the GitHub issues system and GitHub Discussions for reporting bugs, enhancement requests or general questions.
Citing CellExplorer in research and publications
Peter C. Petersen, Joshua H. Siegle, Nicholas A. Steinmetz, Sara Mahallati, György Buzsáki. CellExplorer: A framework for visualizing and characterizing single neurons. Neuron, September 29, 2021; doi: https://doi.org/10.1016/j.neuron.2021.09.002.
Video demonstrating the user-friendly capabilities of CellExplorer
The video can be streamed on YouTube in 4K and is available for download (60MB). For best viewing experience on YouTube, select highest resolution and maximize the video.
Funding
CellExplorer is funded through the Oxytocin U19 BRAIN Initiative Grant, the Lundbeck Foundation, and the Independent Research Fund Denmark.


