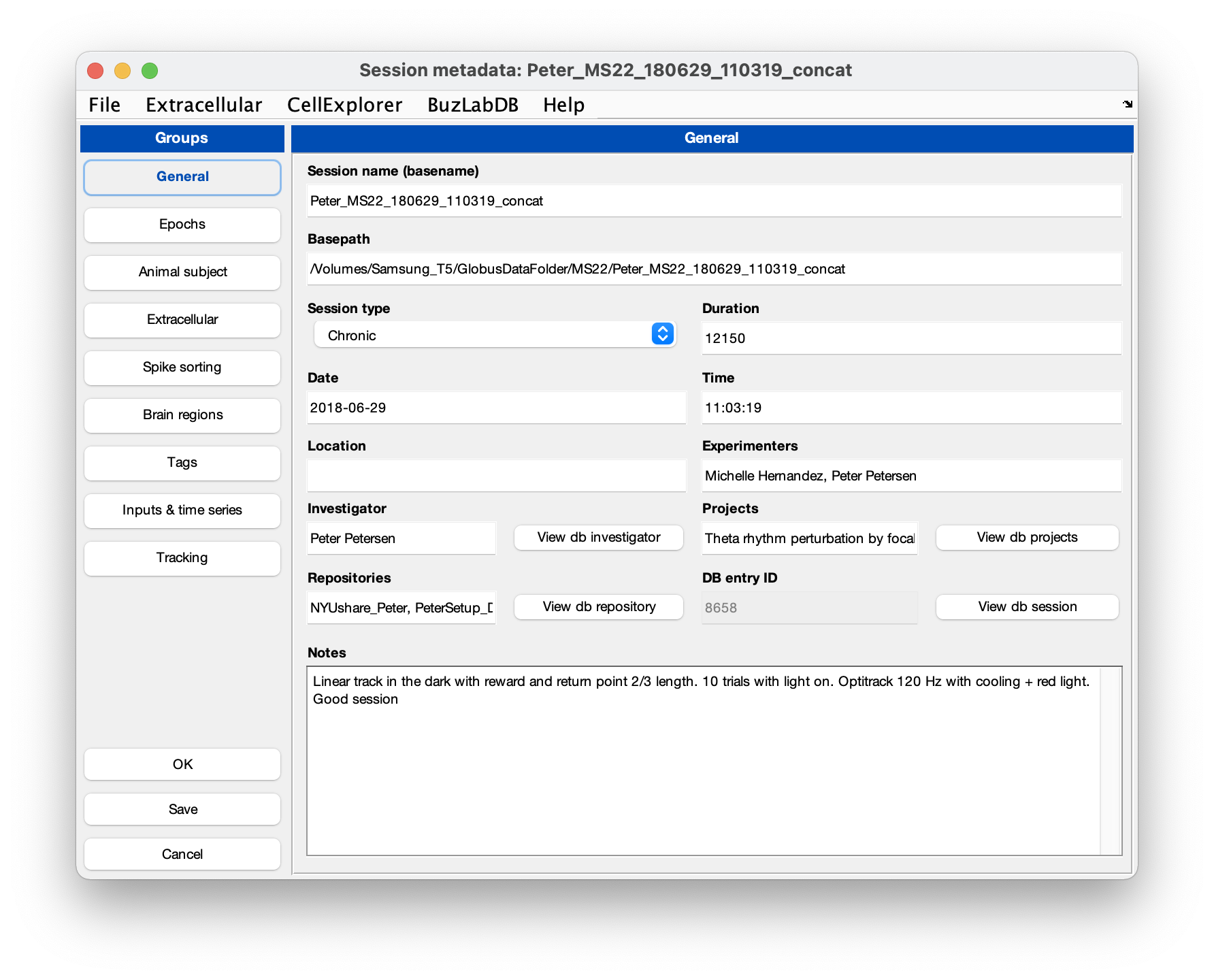
GUI for session metadata
The session GUI allows you to view and manually enter session level metadata through a user-friendly GUI. The interface follows the structure of the session metadata struct, described here, with a tab for each of the main fields of the session metadata struct. Use the left side panel to navigate the tabs.
Table of contents
Side menu
The side menu is organized accordingly to fields in the session metadata struct:
- General: general information about the session like name, date, time, location and experimenters.
- Epochs: temporal aspects of the recording, describing the state of the animal through its environment, the task it is meant to perform and any manipulations.
- Animal subject: contains animal level metadata, including its name, species, strain, and actions performed on the animal. New entries can be added, and existing entries can be duplicated, edited, and deleted.
- Probe implants: allows you to specify any probe implants by the probe design, its implant coordinates, brain region, and orientation
- Optic fiber implants: allows you to specify each optic fiber implant by the optic fiber, its implant coordinates, brain region.
- Surgeries: allows you to specify surgeries performed on the animal by the date, time, place, weight, location, anesthetics, analgesics and antibiotics.
- Virus injections: allows you to specify virus injections by its virus, injection schema, volume and injection rate, brain region and coordinates.
- Extracellular: contains the metadata describing the extracellular data including the number of channels, sampling rate, precision, equipment and electrode groups and layout.
- Spike sorting: Information about spike sorting, including sorting algorithm and format.
- Brain regions: Allen Institute brain regions defined for each extracellular electrodes.
- Inputs and time series: contains description of inputs and time series data.
- Behavioral tracking: describes any behavioral tracking, e.g. video files or types other behavioral tracking data.
Open the session metadata GUI
In Matlab go to the basepath of the session you want to open. Now run the session metadata GUI:
session = gui_session;
The script will detect and load basename.session.mat in the folder. If it is missing, it will show a dialog allowing you to generate the metadata Matlab struct using the template script sessionTemplate. The template script will detect and import metadata from:
- An existing
basename.xmlfile (NeuroSuite) - From Intan’s
info.rhdfile - From KiloSort’s
rez.matfile - From a
basename.sessionInfo.mat(Buzcode) file.
Once you have the sessions struct in the Matlab Workspace you can specify the session struct when opening gui_session:
session = gui_session(session);
You can also provide a basepath as a input:
session = gui_session(basepath);
Compiled versions of gui_session
gui_session can be compiled to a gui_session.exe and a gui_session.app for Windows and Mac respectively. The compiled versions can be used without a Matlab license and without having Matlab installed, but they can also be used independently on a system with Matlab. If Matlab is installed on your system you only need the application (gui_session.exe or gui_session.app), but if you want to use it on a system without Matlab, you have to use the installer (see the gui_session_Installer_web file included with the zip files below).
You can download compiled versions of gui_session for Windows and Mac. The compiled versions are not necessarily the latest version of gui_session.