Opto-tagging tutorial
This tutorial will guide you through the process of tagging your cells by assigning groundTruthClassification-tags to your data in CellExplorer. If you have data that you are interested in sharing please contact us. You can push ground truth cells back to the CellExplorer GitHub repository, or you can send your data by email to us, see instructions below. Cells assigned as ground truth will have a classification label in cell_metrics.groundTruthClassification. Opto-tagged/ground truth cells can be assigned to one or more groups.
Table of contents
- Add your opto-tagged cells to the
cell_metricsstruct - Tagging your cells in CellExplorer
- Saving opto-tagged/ground truth cells to groundTruth folder
- Submit your cells to CellExplorer repository
Add your opto-tagged cells to the cell_metrics struct
Opto-tagged/ground truth cells can be assigned to one or more groups in cell_metrics.groundTruthClassification. If you already have determined the identity of your cells you can simply add them to cell_metrics.groundTruthClassification:
cellIDs_optoTagged = [1,5,10]; % IDs (UIDs) of the opto-tagged cells
name_optoTagged = 'PV_pos'; % Cell line name
cell_metrics.groundTruthClassification.(name_optoTagged) = cellIDs_optoTagged;
Or:
cell_metrics.groundTruthClassification.PV_pos = [1,5,10];
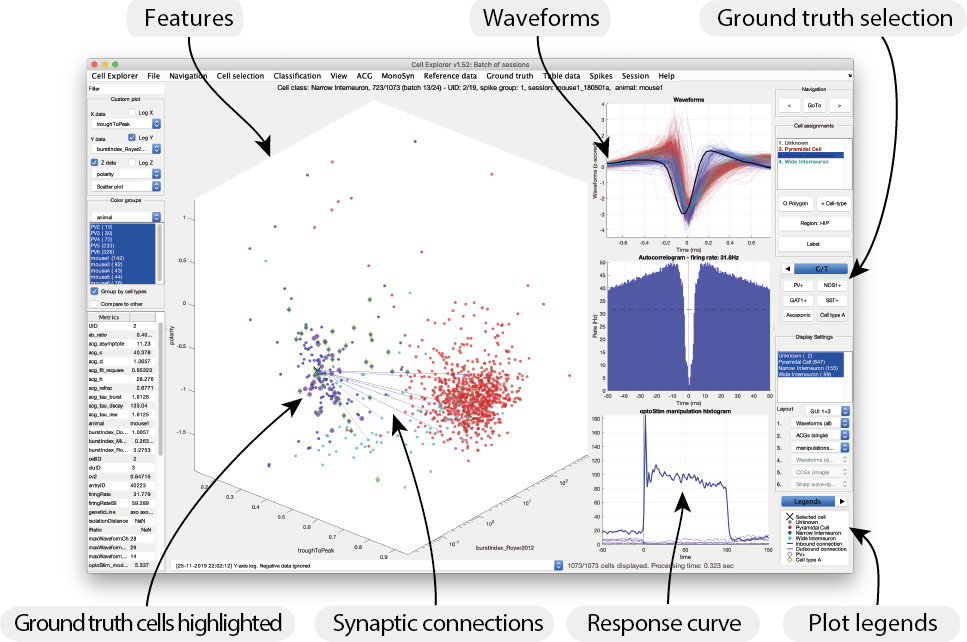
Tagging your cells in CellExplorer
- Launch CellExplorer.
- Activate the manual curation of ground truth classification from the top menu
Ground truth->Perform ground truth cell type classification in current session(s). This opens a tab group in the Cell Assignment tab menu titledG/Tin the right side-panel. - Adjust the highlighted cells using the menu option
Group data->Open group data dialog. This dialog allows you to define how to visualize your tagged cells. - Assign the ground truth tag label to your cells. You can add more tags in CellExplorer and in the preference file
preferences_CellExplorer.m. Each cell can have one or more ground truth classification tags assigned. - Once complete, save the session using the top menu
File->Save.
Saving opto-tagged/ground truth cells to groundTruth folder
Following the process described in the previous sections on tagging your cells, you can submit your opto-tagged cells to the groundTruth folder through the CellExplorer UI, allowing you to share your cells and use them across sessions.
- Launch CellExplorer.
- From the
Ground truthtop menu selectSave tagging to groundTruthData folderto submit your ground truth cells to the+groundTruthData/data folder (centralized ground truth data). The files in the+groundTruthData/folder is organized by sessions.
Submit your cells to CellExplorer repository
You can submit your cells to the CellExplorer repository such that other people can take advantage of your ground truth cells. This allows the community to share their tagged cells such that others can researchers can benefit.
- Push metrics back to main branch of the CellExplorer Github repository: If you cloned or forked the CellExplorer Github repository, you can submit a pull-request to the main CellExplorer branch. We will verify your data and submit then to the main branch of CellExplorer.
- Email metrics to us: You can email your ground truth cells to us at petersen.peter@gmail.com. Provide the
cell_metricsfiles saved to+groundTruthData/. The ground truth cells are organized by sessions, where the files in+groundTruthData/only contains cells with groundTruthClassification from the original session. We will verify your data and submit them to the main branch of CellExplorer.